Section 3 Lab Resources Overview
Here is a very general overview of lab-wide resources that everyone in both sides of the lab should be familiar with. (Some projects also use specific tools that should be separately documented.) As one broad principle to keep in mind: research coordinators in the lab typically stay 2-3 years, but many of our projects span a much longer period of time. We need everyone to perform protocols consistently, and to document their work carefully, so that the people who follow you can continue your work.
3.1 Slack
Slack is the primary method of communication within the lab. You can customize your notification preferences (particularly important on the mobile app) so you are notified of important messages but also are not overwhelmed. Use the ‘@’ feature to get my attention if I need to get involved in a conversation, and make sure that you’re subscribed to the right channels. Key channels for everyone are #general and #random. For our new big lab projects in 2025 we have work channels set up for #decisions-for-others (a continuation of a channel previously labeled #hillblom and then #branch for online studies of decision-making in healthy older adults), #neurotech (now focused mainly on HCD Neurotech grant funded in 2024, but with some residual wrap-up work on our previous NIH-funded neuroethics grants), and #personal-values. Valerie’s been hosting a rich set of readings with discussion at #disability-studies, and a channel for #code has been used mostly for conversations about R and RStudio. More or less in the archives are channels for older lab projects: #caregivers, #dma, and #gbd. We also have channels for non-work-related conversation at #decisionzbop (a channel for music conversation), #food-opinions, #im_upset (originally started early in the COVID-19 pandemic when that topic overwhelmed all our other channels, now a general hub for concern about the state of the world), and #social for informal non-work gatherings and conversations that don’t involve me directly. As a matter of lab policy, we do not have private/locked Slack channels.
Two important caveats about Slack:
- Slack is not considered secure or HIPAA-compliant. No PHI (protected health information) belongs there, and in general any discussion involving individual research participants should be conducted via SECURE: e-mail rather than Slack.
- Slack is not an archive. We can search in our lab account, but it’s not that easy to go back and reconstruct details of conversations and decisions made. So we use Slack for rapid, concurrent conversation–but anything that we might want to refer to later needs to be documented, most likely in GitHub.
3.2 Lab website
Our lab website is one place where we communicate what we do and share our work with the outside world. This is hosted on our public GitHub repository, which you will need a GitHub account to edit. See the README at github.com/DecisionLabUCSF/decisionlabucsf.github.io to learn how to add your team profile. Because this is a public repository, anyone can read the code that we use to build the website, and it also has an open license inviting other investigators to use this code to design their own websites if they wish.
As I say in the README, you can think of GitHub as like a supercharged Google Docs for code, allowing multiple people to work on code together, keeping track of who made what changes and when, making it easy to reverse changes that have been made, and allowing different versions to be developed at the same time. Tools like this are commonly used by software companies to maintain quality and avoid/fix bugs. We won’t be using all of the sophisticated tools in GitHub that they use, but generally I think scientists have a lot to learn from software developers: Facebook and Google can’t survive with sloppy code and neither can we.
3.3 Private Github repo
In addition to our public repository, we also have a private repository that only lab members can view, which is where we document and edit our code, and maintain electronic lab notebooks. See the README at github.com/UCSFMemoryAndAging/decisionlab for details once your account is fully set up with permissions. A basic way of working with GitHub is just editing files on the GitHub website. However, to use GitHub for code that can be utilized by statistical/analytic packages like R and MATLAB, you will need to synchronize the lab repo to a folder on your computer (like a more technical version of Dropbox or Box). For more details on how to do this, see Section 9 of this lab handbook, RStudio Analysis Pathway.
Here are some key initial points:
- As with Slack, no protected data. PIDNs and other assigned identifiers can pop up infrequently, but we should avoid other participant-level data, and especially PHI and participant identifiers (names, dates of birth, addresses, demographics, etc.)
- GitHub works best with text-editable files: code, markdown files, HTML, notebooks and the like. GitHub is much less useful for big files like images, Word and Powerpoint files, and they tend to clog things up for everyone else who is syncing to the repo. Images should be reduced in size whenever possible, and Microsoft Office files (Word, Powerpoint) should stay out of GitHub and go on the R: drive somewhere.
- Please be consistent about naming and file organization, and be obsessive about documentation and comments! Again, other people will pick your work up after you, so please be kind to them in advance. (Quick note that things can get messy if files or folders get renamed after they’re in use, so it’s good to be thoughtful and pick good names for things at the outset–see more about this in the README for the repo.)
- Try to keep line length to about 80 characters, and commit-pull-push!
3.4 R: drive (R:\groups\chiong)
To recap some points from above: Slack can’t be used for protected data or discussion of individual research participants, and isn’t an archive. GitHub is an archive for code and documentation, but also shouldn’t be used for protected data or identifiers (and documentation about individual participants should be limited), and shouldn’t be used for large files such as Microsoft Office documents. So: for our quantitative projects we use the R: drive for large datasets, especially those including personal identifiers, and other useful resources that are too large to put in GitHub. (Quick personal note: I’m often working from my laptop and home computer, on which I only mount the R: drive intermittently–so if you need me to look at something that doesn’t involve PHI, it will often be a lot faster if it lives in GitHub or is temporarily available in Slack.) For instructions on how to mount the R: drive on your computer, see the Technology section of MACipedia.
For using statistical packages, a model for how to utilize the R: drive with the GitHub repo is described in Section 9: RStudio Analysis Pathway). As a general overview, the pathway is:
- Clone the decisionlab GitHub repo to your computer.
- Save original dataset (e.g., a .csv or Excel file from Qualtrics or E-Prime) on the R: drive, and never write to this file again.
- Clean the data, using a script and a logging file that are saved in GitHub, creating a cleaned data file that is saved back on the R: drive.
- Analyze the data using other script and logging files that are saved in GitHub, generating new data files and graphics as necessary that are saved on the R: drive.
Since everyone is using the R: drive for a variety of projects (which will continue on after you leave), keeping our group folder organized is a challenge! Please refer to Guide.docx on the R: drive which explains how the subfolders are organized.
3.5 Box
In our qualitative projects we are now using Box, which is better for involving non-MAC colleagues like in MCL (Medical Cultures Lab) and non-UCSF collaborators. Note that there’s one way in which Box isn’t as good for data security as the R: drive, in that if people have Box Sync enabled then they’ll be carrying around copies of all of the data for a given project on their own laptops. (This is why drive encryption is important, but even so I’ve decided that I’m just not going to think too much about all of these copies of our data wandering around the world in people’s personal devices.) For all of our data projects, we should regularly review who including our collaborators has access to a given folder, and whether such access remains necessary or justified.
3.6 REDCap
Somewhat new for 2025, we are now using REDCap for tracking participants in our studies, including enrollment and consent, and the completion of study activities. REDCap is a very useful tool though it has a bit of a learning curve, in part reflecting some advanced capabilities (used for running things like multi-site randomized clinical trials) and options that go beyond our anticipated uses. It has very good resources for securing data and tracking who should have access to which data, though we’ve heard from other researchers that coordination across site (e.g., with non-UCSF collaborators) can get messy with things like permissions. We’re still learning how to use it, though I’m enthusiastic about it and have found it to be a big improvement so far over what we were doing before.
3.7 Open Science Framework
“Open science” and our lab’s relationship to this idea are discussed in detail in Section 8, Reliability and Open Science. We use OSF to host preregistrations of our studies (data collection and analytic plans that we try to write before we begin a study, and which other researchers can check against our work later on) as well as public datasets and code. To avoid concerns about access and permissions, we’ve been posting all of these materials on my own account, even when they are authored by (and credited to) other lab members.
Organization in OSF is like a Russian doll, with components nested inside of larger components nested inside of still larger components. At the top level is the user’s account, which can contain a number of different projects. Inside each project could be a number of smaller subprojects, which each might represent some piece of the larger project such as the work intended to result in a particular planned publication:
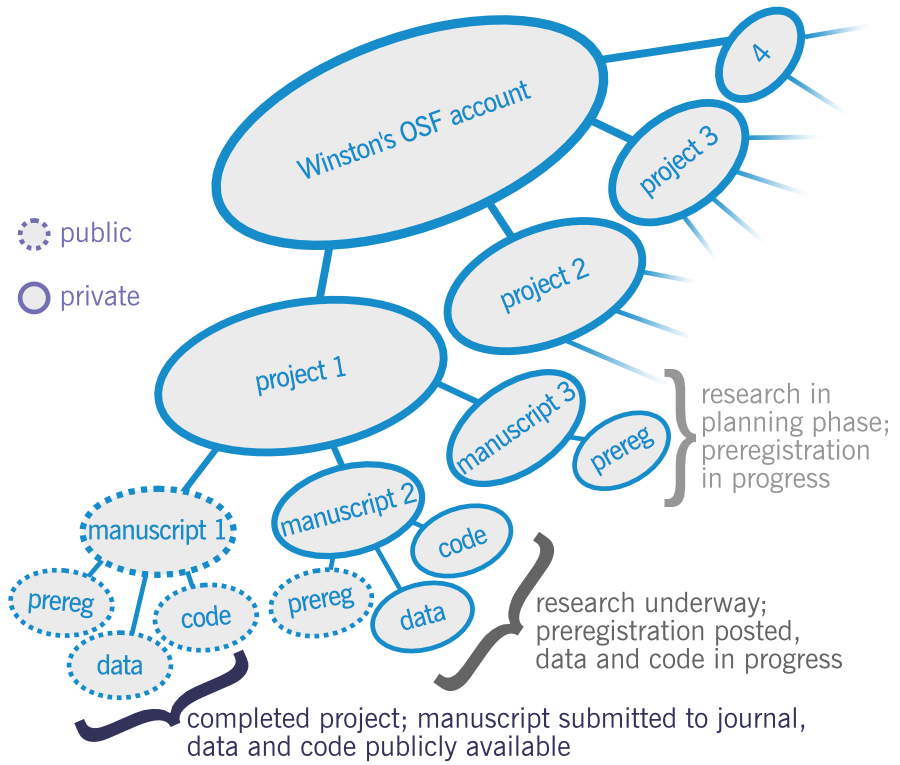
A key distinction in OSF is between what’s “public,” meaning anyone in the world can see it, and what’s “private.” For many projects, this might proceed in different stages, where things start out as private and we gradually open them up to public. It’s useful to note that in OSF, a private component can have public subcomponents inside of it, but a public component can’t have private subcomponents. In this schematic, there’s a private “project 1” that has three components corresponding to three planned publications that are in different stages of completion. For manuscript 1, it’s already complete and has been submitted to a journal. As the work is finished, everything is public: the preregistration, the data, and the code. For manuscript 2, we’ve already planned out the data collection and analysis plans, and so we’ve posted a public preregistration of the project that anyone can read. However, since we’re still working on it, we haven’t made the data or code public yet. For manuscript 3, we’re still planning the data collection and analysis plans, so there’s a private draft registration that hasn’t been posted and made public yet (and the data and code don’t yet exist.)
Currently we have a few big “Projects” in OSF that are private, corresponding to our previous set of big lab projects listed on our website: Neurotech (in “Population survey panel studies”), DMA (“Decision-making in Alzheimer’s disease and related dementias”) and BRANCH (“Online studies of decision-making in healthy aging”). (We preregistered GBD before we came up with this organization system, so the GBD projects are scattered in their own folders.) I’ll be creating some new projects for our new 2024 grants soon.
If you are writing a preregistration for the first time, consider looking at examples such as Wisdom and fluid intelligence in older adults by Cutter Lindbergh and Utilitarian moral reasoning in times of a global health crisis by Rea Antoniou.
3.8 Other useful MAC tools
For these, you’ll generally need to be accessing from campus or, if off campus, signed in through the UCSF VPN (check out it.ucsf.edu/services/vpn for how to get the VPN set up).
3.8.1 LAVA and LAVA Query 5
LAVA is the MAC’s primary database for all research-related patient and participant information, e.g. demographics, visits and scheduling, research diagnoses, specimens.
For our current projects, we use LAVA for the following:
- Getting participants’ contact information to call/email them about enrolling in one of our studies
- Enrolling participants under the ‘Enrollment’ and ‘Scheduling’ tabs once we have seen them for a study
- Utilizing the Contact Log when reaching out to participants in our studies, so that our communication is coordianted with MAC parent projects and with other subprojects
- Looking up participants’ most recent (and past) diagnoses for tracking enrollment numbers or running data analysis
LAVA Query 5 is a (fairly) user-friendly querying tool that allows you to pull information from a selected subgroup of (or all) participants from the main LAVA dataset. You can access this on the front page of the LAVA website.
If you will be running any kind of analysis, you’ll most likely use LAVA Query 5 to pull relevant participant information, download it as a csv file, and merge with your study data files.
For more info how to do this, visit the LAVA Query Protocol and the Pulling LAVA Queries sections of MACipedia. It’s also useful to “ϟ Download Current Data Dictionary” from the front page of LAVA Query 5 (under Data Objects).
3.8.2 MACipedia
As referenced a few places above already, MACipedia has a wealth of information on training and documentation for the MAC. Worth exploring, though can be a bit of a maze to find what you are looking for.
3.8.3 Brainsight/DASH
Powerful and relatively user-friendly tools for visualization of MAC data. Includes individual-subject level graphic summaries of neuropsychological data and viewable neuroimaging data (including “raw” neuroimaging data, for which you will generally want to view the T1 images for neuroanatomy; and W-maps comparing atrophy to age- and sex-matched norms); also with some tools for performing group-level voxel-based morphometric studies that we can talk about separately if you will be running such analyses.
3.9 Deprecated/discouraged tools
Excel. In general, Microsoft Excel is not an appropriate tool for data
analysis or manipulations that are intended for presentation or publication. It
can be used to explore datasets, but usually this is best done within a proper
statistical/analytic software package such as R, Stata or Matlab. Best estimates
are that between 2% and 5% of Excel formulas have uncorrected coding errors, so
if you are using a worksheet with at least 35 formulas then it probably has at
least one error you don’t know about (and if you’re not particularly expert in
using Excel, probably many more). Field studies have shown that almost all
large spreadsheets in the corporate world have multiple errors. If capitalism
supposedly works by efficiently utilizing information to optimally allocate
resources where they can be best used, then someone please explain
to me why businesses still install this program on their employees’ desktops.
In 2012, JPMorgan Chase lost $6.2 billion by relying on a risk model coded in Excel that included a simple formula calculation error. As a commentator noted about weaknesses of reliance on Excel, “There is no way to trace where your data come from, there’s no audit trail (so you can overtype numbers and not know it), and there’s no easy way to test spreadsheets.” This statement provides a decent outline of software requirements for a minimally reliable data system. We should be able to trace where the data that generate our results came from. We should have a record of changes that are made to a file, so we can diagnose if errors are introduced accidentally. (In addition, we need systems in which it’s not so easy to accidentally make changes, such as by distinguishing between viewing and editing; you ought to be able to view a record without worrying that accidentally touching your keyboard will overwrite your data.) And we should have systems in which it’s easy to validate and find out if something has gone wrong.
In the past, we’ve used Excel sometimes to create or input data (e.g., for tracking participants) that is then imported to stats packages. For now, my very strong preference is to do any such work in REDCap since the potential for introduced errors is so high in Excel. Finally, some of the data tools we use (REDCap, Qualtrics [in older projects], LAVA Query) export csv and xls files–our general practice is to save them somewhere on the R: drive and avoid ever touching or rewriting to them directly, only importing from saved files into a proper stats package.
Google Docs. In general, we have found that Google Docs is useful for sharing short-term projects, such as an abstract or IRB submission that needs to be edited by multiple people before submission. However, it has proven to be unwieldy for archiving/storage/documentation–we’ve noted problems with moving/deleting files, with governing permissions and file access, and in general with organization. So anything that we might want to refer back to in a matter of months or years should be appropriately organized in GitHub or the R: drive.